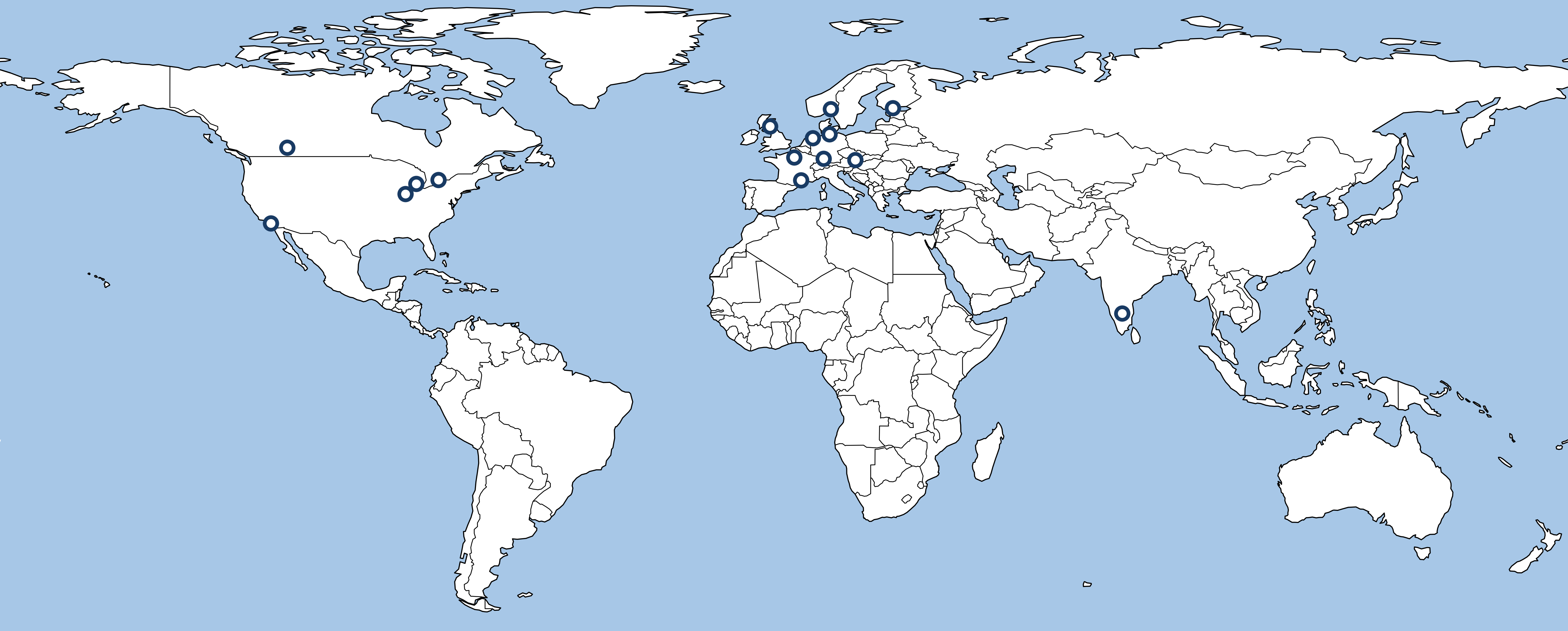
We are a group of academics who investigate polygenic adaptation using experimental and theoretical approaches. Our members are based in ten countries, Germany, Austria, France, Norway, the Netherlands, Canada, Finland, India, the United Kingdom, and the United States, and are at various stages in their academic careers, including postdoctoral, early-career, and advanced. The names and affiliations of our members are listed below in alphabetical order.

Neda Barghi
Max Planck Institute for Evolutionary Biology Plön, Germany
Research area: In our group, we investigate how populations adapt to new environments. We study the genetic basis of adaptation and the effect of different factors such as genetic redundancy, selection strength, and pleiotropy on the adaptive responses. We use experimental and computational approaches such as evolution experiments in Drosophila, high throughput genomic and phenotypic data, and computer simulations.
Research organisms and approaches:Drosophila melanogaster and D. simulans. Experimental and computational.
Lab website: https://www.evolbio.mpg.de/3792274/Max-Planck-Research-Group-Evolution-of-polygenic-traits and https://barghilab.com/
E-mail: barghi@evolbio.mpg.de
Bluesky: @nedabarghi.bsky.social

Yingguang Frank Chan
University of Groningen,
Groningen, Netherlands

Luis-Miguel Chevin
Center for Evolutionary and Functional and Ecology (CEFE), Montpellier, France
Research area: We are interested in adaptation to changing environments in the broad sense, from the genetics of and selection on adaptative traits to the evolution of plasticity, and eco-evolutionary dynamics. Work most related to the network involves the role of pleiotropy in parallel evolution, the evolution of genetic correlations between traits at mutation-selection balance, or more recently the evolution of chromosomal fusions under epistatic selection.
Research organisms and approaches: Theory, experiments with the halotolerant microalgae Dunaliella salina, analyses natural populations monitored over long-term (mostly birds). Population and quantitative genetics.
Lab website: https://www.cefe.cnrs.fr/fr/component/content/article/474-luis-miguel-chevin
and https://lmchevin.weebly.com/
E-mail: luis-miguel.chevin@cefe.cnrs.fr
Bluesky: @lmchev.bsky.social

Maud Fagny
GQE – Le Moulon, Université Paris-Saclay – CNRS – INRAE – AgroParisTech, France
Research area: Crops yield a set of more or less correlated, polygenic traits, governed by complex gene regulatory networks (GRN) with epistatsis and pleiotropy. They depend on both the genetic background of the crops and the environment. We investigate how the structure of GRN impact the response and adaptation of crops to their environment. We develop three research axes: (i) investigating the rewiring of crops GRN in response to abiotic stresses; (ii) assessing the influence of genetic variations on GRN rewiring and investigating the interaction between GRN structure and polygenic selection on crops adaptation; (iii) use GRN information to improve crop yield prediction models in response to climate change.
Research organisms and approaches: We perform both high-throughput molecular data analyses using quantitative genomics and systems biology (gene regulatory network inference) approaches, and develop more theoretical population genetics models. Our model organism is maize response to drought.
Lab website: https://moulon.inrae.fr/en/personnes/mfagny/ and http://maud.fagny.fr
E-mail: maud.fagny@inrae.fr
Bluesky: @maudfagny.bsky.social

Frédéric Guillaume
University of Helsinki, Finland
Research area: We study the genetics of adaptation both in vivo, with an experimental evolution approach using the red flour beetle Tribolium castaneum, and in silico, with genetically explicit forward-time simulations with our software Nemo. We focus on integrating quantitative and population genetics approaches to understand how genes covary in either allele frequency changes over time or gene expression levels in different selective environments to decipher the genetic basis of fitness and gene expression changes. In silico experiments allow us to generate genomic and phenotypic data to test the statistical power of new approaches to detect genomic signals of selection or to test hypotheses related to how the structure of the genotype-phenotype-fitness map affects things like the rate of adaptation to new environments.
Research organisms and approaches: Experimental evolution with Tribolium castaneum. Computational approach with Nemo, a forward-time, genetically and spatially explicit individual-based simulation program.
Lab website: https://www.helsinki.fi/en/researchgroups/eco-evolutionary-dynamics
E-mail: frederic.guillaume@helsinki.fi
Bluesky: @fredguillaume.bsky.social

Angela M. Hancock
Molecular Basis of Adaptation Lab
Purdue University, USA
Research area: My research aims to clarify how complex traits evolve. We integrate computational approaches with lab-based experimental work to characterize the polygenic basis of adaptation, and identify the loci and variants involved in quantitative trait variation and reconstruct their evolutionary histories.
Research organisms and approaches: Plants including Arabidopsis thaliana, Experimental
Lab website: https://ag.purdue.edu/directory/mahancoc
E-mail: ahancock@purdue.edu
Bluesky: @ahancock.bsky.social

Thomas F. Hansen
University of Oslo, Norway
Research area: I am a theoretical biologist interested in conceptual, philosophical, historical and methodological aspects of biology in general and evolutionary biology specifically. I have my main technical expertise in theoretical population genetics and evolutionary quantitative genetics. My main lines of research concern the potential for adaptation (evolvability), the representation and measurement of evolvability, selection, adaptation and rates of evolution, the study of adaptation with phylogenetic comparative methods, the evolution of genetic architecture, and the relationship between micro- and macroevolution.
Research organisms and approaches: My work is predominantly theoretical, but I often collaborate with empiricists and in this way I have experience with a variety of study organisms.
E-mail: thomasha@uio.no

Matthew Hartfield
Institute of Ecology and Evolution, The University of Edinburgh, UK
Research area: My lab investigates the interaction between how a species reproduces, and how it interacts with genetic selection. To this end we are investigating how low recombination and uniparental reproduction (e.g., via self-fertilisation) influences the selective response of polygenic traits. We are tackling these questions through creating novel mathematical models and analysing genomes from self-fertilising species.
Research organisms and approaches: Theoretical population genetics; genomics; bioinformatics. Study organisms currently include Caenorhabditis elegans and Arabis alpina.
Lab website: hartfieldlab.com
E-mail: m.hartfield@ed.ac.uk

Laura K. Hayward
Institute of Science and Technology Austria

Joachim Hermisson
Mathematics and BioSciences Group (MaBS)
University of Vienna, Austria
Research Area: Theoretical population genetics, mathematical biology. Process and patterns of adaptation: influence of the genotype-phenotype map, linkage and epistasis, population structure, demography, fluctuating selection, molecular footprints.
Research organisms and approaches: Modelling, theory
Lab website: http://www.mabs.at
E-mail: joachim.hermisson@univie.ac.at

Kavita Jain
J Nehru Centre for Advanced Scientific Research, Bangalore, India
Research area: Currently, I am working on understanding the joint effect of random genetic drift, recombination and stabilizing selection on the steady state properties and the adaptation dynamics of a population in the weak mutation regime.
Research organisms and approaches: Theoretical
Lab website: https://www.jncasr.ac.in/jain/
E-mail: jain@jncasr.ac.in

Emily Josephs
Michigan State University, USA
Research area: The Josephs lab is broadly interested in understanding how various evolutionary forces, like drift and selection, shape patterns of genetic variation and trait variation in plant species. We tackle these questions using population genomic and quantitative genetic techniques with a focus on how these quantitative (complex) traits change across environments because of phenotypic plasticity. We also investigate how different types of genomic changes, like transposable elements and polyploidization, along with changes in sexual systems and dispersal patterns, shape quantitative trait variation and the ability of plants to respond to selection.
Research organisms and approaches: Capsella bursa-pastoris (Shepherd’s purse), Arabidopsis thaliana, Chamaecrista fasciculata, members of Boechera and Borodinia. Mostly experimental approaches
Lab website: Josephslab.gitbhub.io
E-mail: josep993@msu.edu
Bluesky: @emjo.bsky.social

Anthony D. Long
Department of Ecology and Evolutionary Biology, University of California, Irvine, USA

Luisa F. Pallares
Evolutionary Genomics of Complex Traits group
Friedrich Miescher Laboratory of the Max Planck SocietyMax Planck Institute, Tübingen, Germany
Research area: Our research focuses on understanding (a) the genetic basis of between-individual variation in complex traits and (b) how such complex genetic architectures, instead of being static properties of a trait, get re-shaped when populations are exposed to different environments (genotype-by-environment interactions or GxE). In addition, we are particularly interested in (c) understanding how phenotypic robustness is regulated in such traits. That is, we aim to understand not only why individuals in a population look different from each other, but also why some individuals are more vulnerable than others when exposed to perturbations like stressful or new environments.
Research organisms and approaches: Drosophila melanogaster (experimental + theoretical)
Lab website: www.pallareslab.org and www.fml.tuebingen.mpg.de/9004/pallares-lab
E-mail: Luisa.pallares@tuebingen.mpg.de
Bluesky: @luisapallares.bsky.social

Himani Sachdeva
Faculty of Mathematics and Department of Evolutionary Biology, Faculty of Life Sciences, University of Vienna, Austria
Research area: I use mathematical models to study how genetic variation is maintained in natural populations, how populations respond to selection, and the longer-term outcomes (local adaptation, speciation, extinction) that arise from the interplay of evolutionary and demographic processes acting over different spatio-temporal scales. A key goal is to better understand the limits of genomic inference, particularly to what extent we can (or cannot) disentangle the subtle signatures of polygenic selection from those of spatial population structure.
Research organisms and approaches: theoretical and computational
Lab website: https://mathevogen.github.io/index.html
E-mail: himani.sachdeva@univie.ac.at

Christian Schlötterer
Institut für Populationsgenetik
Vetmeduni Vienna, Austria
Research area: My laboratory is using experimental evolution to understand polygenic adaptation. Specifically, we ask how genetic redundancy, pleiotropy, non-additive effects and linkage disequilibrium affect the genomic response during adaptation to new complex environments. Our experimental evolution studies incompass experiments with highly complex founder populations as well as simple founder populations consisting only of two genotypes.
Research organisms and approaches: Our pet organism is Drosophila. While most research is done with D. simulans, we also conduct experimental evolution with D. melanogaster and D. mauritiana.
Lab website: http://www.vetmeduni.ac.at/en/population-genetics/
E-mail: schlotc@gmail.com

Jacqueline Sztepanacz
Department of Ecology and Evolutionary Biology, University of Toronto, Canada
Research area: Research in my is focused on testing evolutionary theories for how genetic variation evolves in populations, and specifically, how genetic correlations caused by pleiotropy affect the distribution of genetic variation across organisms and their evolvability. We are interested in how genetic correlations caused by the shared genome of males and females constrain (or accelerate) the evolution of sex differences, whether pleiotropy can lead to evolutionary constraints that affect extinction risk over macro-evolutionary time, and how genetic variation and selection vary across time and space. We also develop statistical methods for multivariate quantitative genetic studies.
Research organisms and approaches: Drosophila
Lab website: https://sztepanacz.eeb.utoronto.ca
E-mail: j.sztepanacz@utoronto.ca

Diethard Tautz
Max Planck Institute for Evolutionary Biology Plön, Germany

Henrique Teotónio
Experimental Evolutionary Genetics
Institute de Biologie, École Normale Supérieure, Paris Science Lettres Research University, France
Research area: Our focus is investigating how organisms adapt to changing environments, employing experimental evolution techniques with the roundworm nematode Caenorhabditis elegans. We’ve cultivated domesticated C. elegans populations, characterized by their standing genetic diversity, diverse reproductive modes, and varying recombination rates, to study adaptation. Additionally, we’ve developed the CeMEE panel, a collection of recombinant inbred lines, enabling us to map quantitative trait loci (QTL) with unparalleled precision for a metazoan. Our research uses quantitative genetics and population genomics to analyze evolutionary diversification trends in real-time.
Research organisms and approaches: Experimental, C. elegans
Lab website: https://www.ibens.bio.ens.psl.eu/?rubrique28
E-mail: teotonio@bio.ens.psl.eu

Sam Yeaman
Department of Biological Sciences, University of Calgary, Canada
